JOSEPH HOPKINS
Applied Data Scientist
Certified Data Scientist, Level 1

Sepsis Outcomes Newborns in Amhara, Ethiopia
Synergy Research Award
Woodruff Health Sciences Center
Rollins School of Public Health, Nell Hodgson Woodruff School of Nursing
Emory University
Research Background
Sepsis, defined as a systemic bacterial infection, is estimated to be responsible for as much as one quarter of neonatal deaths globally. Approximately 30–40% of infections resulting in neonatal sepsis are transmitted at the time of birth. Early-onset infections are often acquired around the time of delivery through vertical mother-to-infant transmission. Late-onset infections present after delivery (typically after 3 to 7 days of age) and are attributed to organisms acquired from the neonate’s interactions with the hospital, home, or community environment. Low birth weight is an established risk factor for sepsis with preterm, low birth weight infants having a 3–10 times higher incidence compared to full-term normal birth weight infants.
However, little is known about the incidence, risk factors, or etiology of neonatal sepsis in sub-Saharan Africa (SSA). The WHO reports “newborns are at higher risk of acquiring health care-associated infection in developing countries, with infection rates three to twenty times higher than in high-income countries.” A study in Nigeria reported 6.5 cases of neonatal sepsis per 1,000 live births in a referral hospital, and 21 cases of neonatal sepsis per 1,000 live births were reported from a referral hospital in Zimbabwe. There are no population-level estimates of neonatal sepsis in Ethiopia.
The study at hand will be the first known work to rigorously examine the burden of facility-acquired infections among facility-born neonates at healthcare facilities with limited WASH infrastructure and practices in a low-income country.
Source:
WHSC Synergy Award Proposal, Christine L. Moe, PhD & John N. Cranmer, DNP, MPH, MSN, ANP-BC
Data Science Challenge
The primary goals of my work in this research were to describe the (1) incidence and (2) etiologic agents of neonatal sepsis in normal and low birth-weight study subjects. Two specific challenges lent themselves to solutions using data science tools and methods:
-
Data wrangling. The research team gathered data from the summer of 2018 through summer 2019 in support of the research aims defined above. The data were collected on paper forms and later manually entered as digital records. The dataset needed cleaning, validation, and pre-processing in advance of analysis.
-
Modeling. The research proposed analysis of several aspects of the data, including incidence and co-variates (such as birth weight) of neonatal sepsis. I planned to use inferential models as implemented in common data science libraries like
sci-kit learnorstats modelsfor this analysis, even though the purpose was more inferential than predictive.
Data Wrangling
The research team collected data over a one-year period at two healthcare facilities in the Amhara, Ethopia region (Felegehiwot Hospital and Debretabor Hospital). The data were then manually transcribed into a research data capture tool called REDCap and exported to Excel.
Once the data were exported from REDCap, our process included the following steps:
-
De-identification. Because of time constraints, I was not able to complete the necessary training with Emory’s IRB to handle personally identifiable information, so the research team had to de-identify the dataset before I could handle it.
- Splitting. In order to keep the following steps manageable in terms of scope, I split the de-identified data into the following sources, based on the forms the enumerators had filled out in interviews with the study subjects, laboratory results of blood cultures for potentially sick infants, and laboratory results of hand-rinse samples from study participants:
- Background. Unique identifier for the mother; 3 demographic fields describing the municipal subdivisions where the mother resides; administrative information not needed for analysis.
- Baseline. First interview with the mother. Includes extensive mother, father, and household demographic information, along with OB, pregnancy, and birth history.
- Follow-up. Interviews at 7 and 14 days post-partum. Contains clinical information on mother and baby.
- Clinical Sample Results. Contains laboratory results of blood work for children suspected of having sepsis, including which microbes were found and their antimicrobial resistance profile.
- Handrinse Wash Sample Results. This was the only data source with the ID of both the mother and the child. Used exclusively for reconciliation.
-
EDA and Cleaning. Because the data were collected on paper forms and transcribed into REDCap, many data fields contained inconsistent data types and formats that needed cleaning up. Many categorical variables needed to be translated into indicator variables (dummy variables).
-
Reconciliation. The most challenging aspect of the data cleaning was the lack of unique, consistent identifiers for each study subject (mother and chilren). I developed several tools for helping the team in Ethiopia to identify and reconcile these IDs so we could link all the data sources together. Unfortunately, the time needed to complete this task exceeded the time available for the Capstone, so the fully combined dataset was not available for Capstone analysis.
- Visualization. I built several visualizations to help plan for pre-processing and to understand the relationships among key study variables. The visualizations are available in the repository.
Modeling
Because the fully consolidated dataset was not available during the window Capstone window, I fell back to analyzing segments of the dataset. Specifically, I completed the following analyses, which I’ll describe briefly below.
- K Means and DBSCAN clustering
- K Nearest Neighbors and Logistic Regression on birthweight status
All these models were implemented using sci-kit learn.
K-Means and DBSCAN clustering
I hoped unsupervised learning might reveal some latent patterns in the dataset, so I ran both K-Means clustering and DBSCAN models on both the Followup and Clinical Sample data. The best silhouette scores for each model and dataset, along with model hyperparameters follow.
| Dataset | K-Means | DBSCAN |
|---|---|---|
| Follow-up Interviews | 0.87 k=2 |
0.79 epsilon=0.68 4 clusters |
| Etiologic Agents & AMR | 0.36 k=9 |
0.32 epsilon=0.1 5 clusters |
Because so much of the data was either categorical (and had been split into binary indicators) or dichotomous, both clustering techniques failed to reveal anything meaningful. Even in the follow-up interview clustering, where silhouette scores were relatively high, the clusters themselves revealed nothing noteworthy in excess of where the interview happened and whether or not the subject deceased.
Predicting Low Birth Weight
More interesting was classification on an infant’s birthweight status. Each baby in the study was classified as either “normal” birthweight (if their recorded birthweight was greater than or equal to 2,000g) or “low” birthweight (otherwise). Of our 615 observations, 19.4% of subjects were classified as “low birth weight”, so baseline accuracy the model needed to outperform was about 81%.
The Followup interview dataset contained 615 observations, and 156 of those contained at least one null value. I experimented with a Python package called autoimpute to perform Predictive Mean Matching imputation for those records. The imputation proved to be fairly fragile due to non-convergence (throwing run-time errors every time it was run but twice), so I instead omitted columns that contained null values and ran two classification models: K Nearest Neighbors and Cross-validated Logistic Regression.
K Nearest Neighbors
Grid-searching over 18 combinations of hyperparameters resulted in a “best” KNN model with 91% accuracy, noticeably above the baseline accuracy of 81%.
| Actual + | Actual - | |
|---|---|---|
| Pred + | 18 | 1 |
| Pred - | 13 | 122 |
| Metric | Value |
|---|---|
| Baseline Accuracy | 80.65% |
| Model Accuracy | 90.91% |
| Specificity | 99.19% |
| Sensitivity | 58.06% |
Subsequent modeling with logistic regression outperformed this, however, so I did not iterate this model.
Cross-Validated Logistic Regression
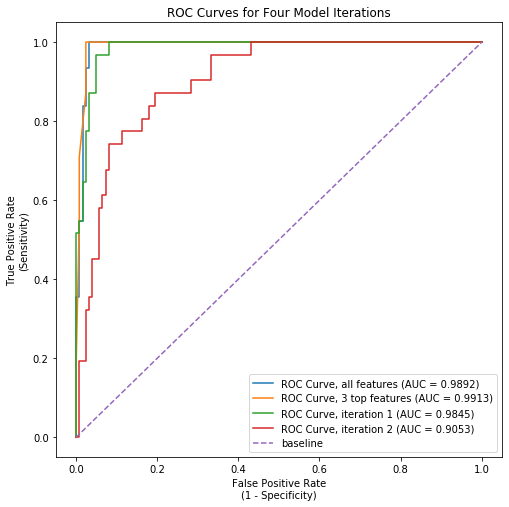 Logistic Regression performed even better than KNN, with an initial model clocking in at 96.8% accuracy. Examining this model more closely revealed that a small subset of features were driving most of the explanatory power of the model. I ran 3 additional iterations that excluded progressively more features. The final model, which used just three features (number of previous low birthweight births, number of previous pre-term births, and number of previous live births), accurately predicted birthweight status over 95% of the time. The AUC-ROC for this model exceeded that of the initial regression model slightly (see image nearby).
Logistic Regression performed even better than KNN, with an initial model clocking in at 96.8% accuracy. Examining this model more closely revealed that a small subset of features were driving most of the explanatory power of the model. I ran 3 additional iterations that excluded progressively more features. The final model, which used just three features (number of previous low birthweight births, number of previous pre-term births, and number of previous live births), accurately predicted birthweight status over 95% of the time. The AUC-ROC for this model exceeded that of the initial regression model slightly (see image nearby).
This could be a valuable tool for predicting birthweight status for non-first-time mothers. It’s worth noting that because we did not yet have a fully consolidated dataset, this finding – while promising – is only preliminary.
The confusion matrix on the test dataset (n=154) and summary model metrics follow.
| Actual + | Actual - | |
|---|---|---|
| Pred + | 120 | 3 |
| Pred - | 4 | 27 |
| Metric | Value |
|---|---|
| Baseline Accuracy | 80.65% |
| Model Accuracy | 95.45% |
| Specificity | 97.56% |
| Sensitivity | 87.10% |